GeneHunter
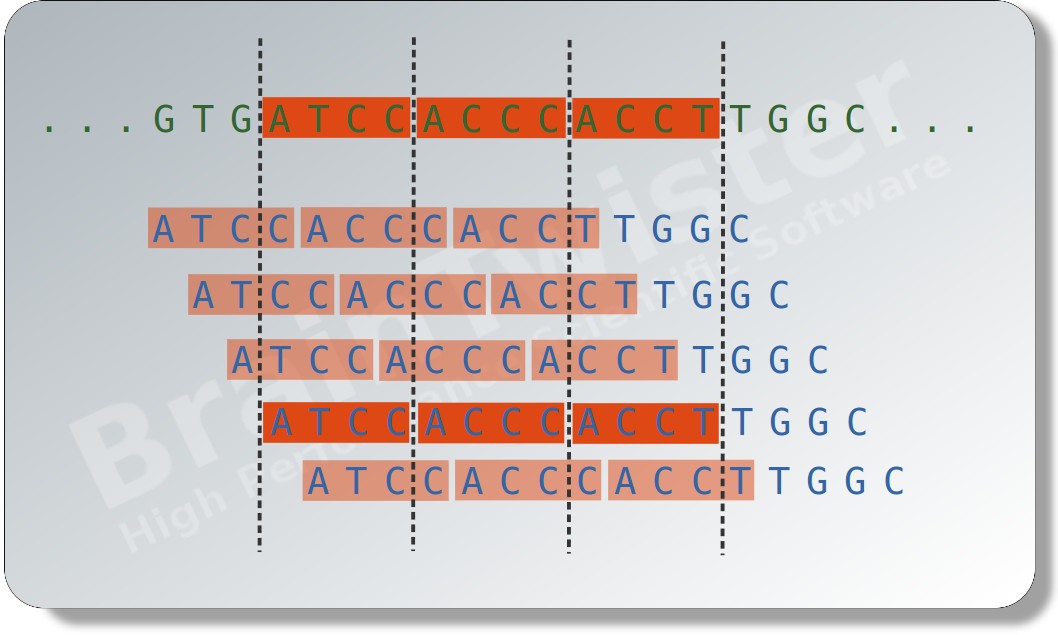
A hash map based algorithm with fixed sized sequence tokens is used to locate matching regions. Then, the best match is determined by the match quality calculated using the local alignment algorithm of Smith-Waterman. Finally, the organism and gene infomations are extracted from the NCBI taxonomy and GenBank database.
GeneHunter is using the nucleiotide and taxonomy databases of National Center for Biotechnology Information (NCBI) to identify organisms and genes from DNA sequencer data.